Zinc Enzymes
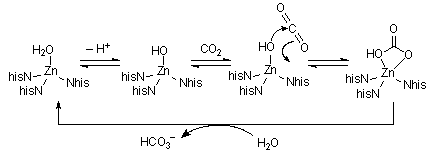
Carbonic Anhydrase| Carbonic anhydrase regulates the dissolution / removal of carbon dioxide
to / from water. It has a mass of about 30kDa, with a zinc ion bound by
3 Nhis and a water molecule. The high local charge dramatically increases
the acidity of this bound water (pK |
|

|
Carboxypeptidase A is the enzyme responsible for the degredation of protein
chains in your pancreas. It has a mass of about 34kDa, and contains one
zinc ion at the active site, bound by 2xNhis, Oglu, and a water molecule.
The water is displaced when a substate binds. A number of other important
protein sidechains (one glutamate and three arginine residues) point into
the active site, and help to orient the peptide chain during hydrolysis.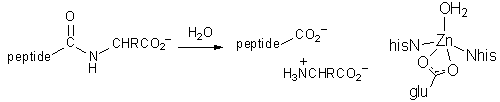 |
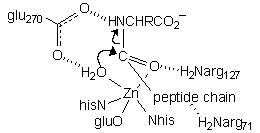
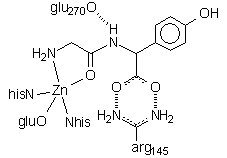 This structure shows the dipeptide GlyTyr bound to the active site. The terminal glycine amino group binds where water would normally be; in a longer peptide the amino nitrogen would be held aside by hydrogen bonding interactions between the chain and both arg71 and arg127. Note the two critical interactions of the tyrosine residue: H-bonds between the C-terminus carboxylate and arg145, an H-bond between the Tyr amino nitrogen and glu270, and the coordination of the carbonyl group to the zinc. These latter two interactions stabilize the amino N as a leaving group and activate the C=O group toward attack by what would be a Zn-bound H2O.
This structure shows the dipeptide GlyTyr bound to the active site. The terminal glycine amino group binds where water would normally be; in a longer peptide the amino nitrogen would be held aside by hydrogen bonding interactions between the chain and both arg71 and arg127. Note the two critical interactions of the tyrosine residue: H-bonds between the C-terminus carboxylate and arg145, an H-bond between the Tyr amino nitrogen and glu270, and the coordination of the carbonyl group to the zinc. These latter two interactions stabilize the amino N as a leaving group and activate the C=O group toward attack by what would be a Zn-bound H2O.
|
|
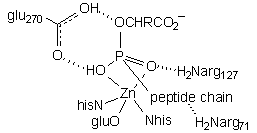
| This structure employs a phosphonate inhibitor, wherein the tetrahedral phosphorus atom (orange) is thought to model the transition state for nucleophilic attack on the carbonyl. The key interactions are: 1) arg145 H-bonding to the C-terminus carboxylate 2) glu270 H-bonding to phosphonate O3 and O2, which would be the cleaved peptide amino N and attacking water in the functional transition state 3) Zn coordinating to phosphonate O2 and O1, which would be the attacking water and peptide carbonyl in the functional transition state 4) arg127 and arg71 H-bonding to phosphonate O1 and the third peptide carbonyl, respectively. |
|
|
|
|
Alcohol dehydrogenase catalyzes the conversion of primary alcohols to
aldehydes, the first step in the metabolism of our society's favourite recreational
drug. This is formally a redox reaction, and uses NAD+ as a cofactor,
although electrons do not pass through the zinc centres. Instead, the Zn
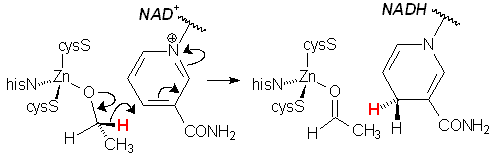
serves only to bind and activate the substrate, acting as a Lewis acid. Mammilian liver alcohol dehydrogenase consists of 2 identical 40kDa subunits, each of which contains two Zn ions. One of these subunits is shown at left. One Zn serves a purely structural role, bound to four Scys groups. The catalytic zinc centre is located in a large binding cleft and is bound by 2 Scys, one Nhis, and a labile H2O, which is replaced by the alcohol substrate. |
| The pocket is surrounded with protein sidechains that bind the NAD+ cofactor via a series of hydrogen bonds (red lines), and orient the face of the nicotinamide ring toward the zinc-bound alkoxide, thereby facilitating transfer of a hydride ion. In this structure, pentafluorobenzyl alcohol is bound to the zinc (but reacts no further due to deactivation by the F groups). Note the juxtaposition of alcohol C1 (H– donor) and NAD+ C4 (H– acceptor) (green line). | |
|
This page is maintained and copyright by W. Stephen McNeil at UBC Okanagan.
All educational works available on this page are licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 2.5 Canada License. ![]()