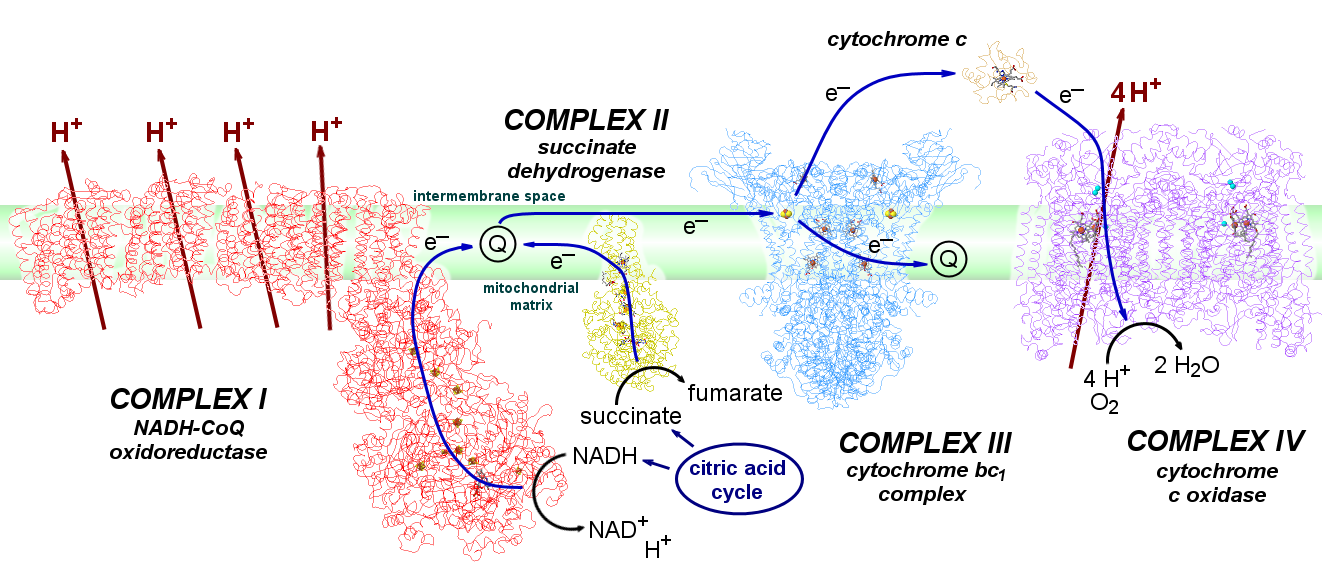
Mitochondrial Electron Transport Chain
A series of metalloproteins bound to the inner membrane of mitochondria carry electrons from the strongly-reducing products of the citric acid cycle (NADH, succinate) to highly-oxidizing O2. The electrons pass through a series of metal clusters and cofactors, and the electron transfer is coupled to the transport of H+ across the membrane, from the interior matrix space of the mitochondria to the intermembrane space, creating a proton concentration gradient. The potential energy of that gradient drives the synthesis of ATP when protons flow back across the membrane through ATP synthase.
The four large proteins associated with mitochondrial electron transport are called Complexes I through IV. Electron transfer between these them is accomplished by the mobile coenzymes ubiquinone (within the lipid membrane, from Complexes I and II to Complex III) and cytochrome c (in the intermembrane space, from Complex III to Complex IV).

| Complex | subunits | MW | redox centres |
| I: NADH-ubiquinone oxidoreductase | 44 | ~980 kDa | FMN, 8 FeS clusters (2x[2Fe2S] and 6x[4Fe4S]) |
| II: succinate dehydrogenase | 4 | 120 kDa | FAD [2Fe2S] [4Fe4s] [3Fe4S] cyt b560 |
| III: cytochrome bc1 complex | 2 x 11 | 480 kDa | cyt b562, cyt b566, Rieske FeS, cyt c1 |
| IV: cytochrome c oxidase | 2 x 13 | 420 kDa | CuA, cyt a, CuB/cyt a3 |
Complex I - NADH-ubiquinone oxidoreductase
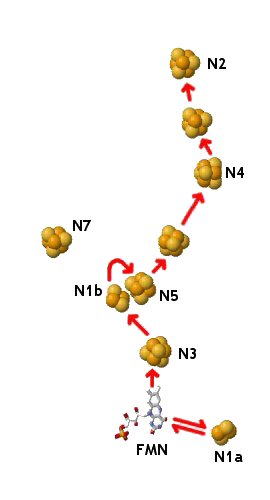
Complex I is by far the largest of these complexes, consisting of >40 protein chains in two domains that form an L-shape -- one hydrophobic domain lies within and is oriented parallel to the membrane, while a hydrophilic arm entends into the mitochondrial matrix. Complex I transports two electrons from NADH in the mitochondrial matrix to Q within the membrane, via FMN and a series of seven iron-sulfur clusters. FMN can break up the simultaneous transfer of two electrons from NADH (along with a proton, transferring as H–) into two separate one-electron steps, as required for electron transfer through the iron-sulfur clusters.
 |
|
|
Structure of hydrophilic domain of Complex I from T. thermophilus, from Sazanov, et al. Science 2006, 311, 1430. PDB 2FUG. |
|
The structures of the complete Complex I from Thermus thermophilus and Escherichia coli are known. The prokaryotic version of the enzyme is far simpler than that found in mitochondria, but analogues of all these subunits are seen in the mitochondrial form, and all the redox centers are equivalent. The hydrophilic domain of Complex I projects into the mitochondiral matrix (or cytoplasm in its prokaryotic forms). In eukaryotes, it has an electron transfer pathway consisting of a bound FMN and eight FeS clusters. (In T. thermophilius and E. coli, a ninth FeS cluster designated N7 lies over 20 Å away from this chain, and is not thought to take part in electron transfer.) Two of the FeS cofactors are [2Fe2S] clusters (N1a and N1b) and the others are [4Fe4S] (N2 through N6). NADH is thought to bind near the FMN. The electron transfer path then passes from FMNH2 through N3, N1b, N5, N6a (or 6b), N4, N6b (or 6a), and N2, which lies less than 10 Å from a proposed quinone binding site. N1a is too far from N3 for direct electron transfer between these clusters, but it may be invovled in preventing the generation of highly-reducing falvosemiquinone FMN–• by temporarily storing the extra electron. That is, FMNH2 could donate two electrons nearly simultaneously -- one to N3 and one to N1a. Once the N3 becomes reoxidized (leading to quinone reduction), N1a can release the extra electron back through FMN back in to the chain.
|
Structure of complete Complex I from T. thermophilus, from Badaran, et al. Nature 2013, 494, 443. PDB 4HEA. |
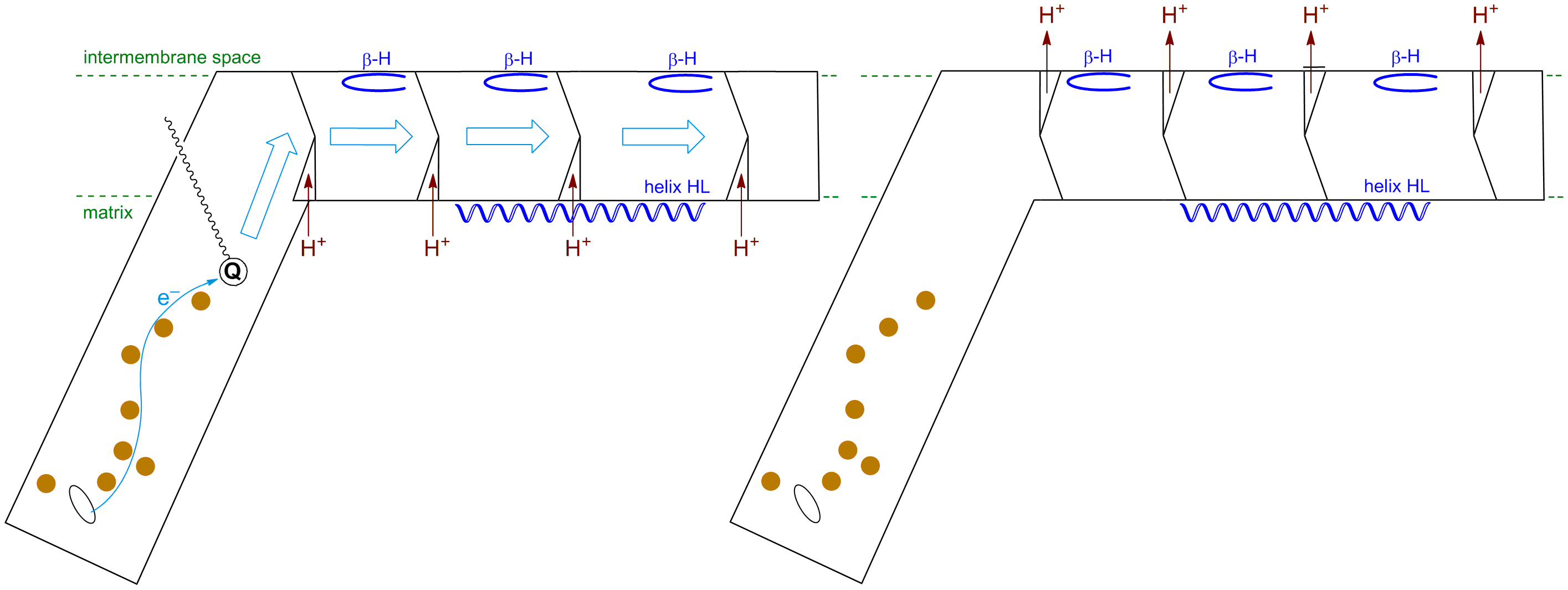
The hydrophobic domain of Complex I lies primarily in the membrane. In the T. thermophilus structure, it spans a distance of 180 Å, and consists of seven subunits and over 60 trans-membrane helices. The ends of these helices are anchored by β-hairpin turns on the equivalent of the intermembrane space side, and by a long helix (HL) running parallel to the membrane surface on the equivalent of the matrix side (shown in blue). The energy associated with the downhill transfer of electrons from NADH to Q through the FeS clusters is used to induce a series of conformational changes in the centre band of the transmembrane helices (blue arrows): proton channels are loaded with H+ from the matrix upon quinone reduction in the binding site, which are then are expelled to the intermembrane space once reduced quinolate dissociates. Thus the electrochemical energy of the redox process is harnessed to pump protons across the membrane.
|
Complex II - Succinate Dehydrogenase
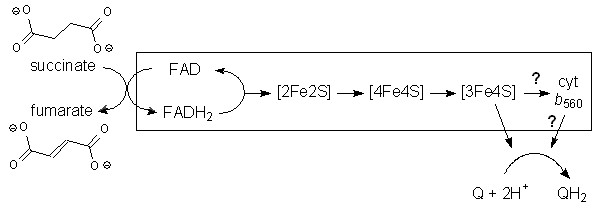
Complex II oxidizes succinate (–O2CCH2CH2CO2–) to fumarate (trans-–O2CCH=CHCO2–) and reduces Q to QH2 within the membrane. The transfer proceeds initially via an FAD cofactor (which, like FMN in Complex I, breaks the 2e reduction into two separate 1e processes), and then through a series of three iron-sulfur clusters. The first is a [2Fe2S] cluster where one of the Fe atoms is bound by an aspartate oxygen rather than the usual cysteine sulfur. The second and third are normal [4Fe4S] and [3Fe4S] clusters. All these groups are in the hydrophilic protein chains external to the membrane (beige). Finally, electrons pass through the heme iron of cytochrome b560 (within the hydrophobic protein inside the membrane, yellow), and then outside the protein to ubiquinone, which binds near the heme group. (Or maybe not. Mutation studies on E. coli Complex II suggest that electron transfer proceeds directly from the [3Fe4S] cluster to Q, bypassing the heme. However, the redox potential of the E. coli b heme is different than that of mitochondrial Complex II, so the precise extent of involvement by the heme isn't wholly clear.)
| Structure of Complex II from Sus scrofa (pig), from Sun, et al. Cell 2005, 121, 1043. PDB 1ZOY. |
|
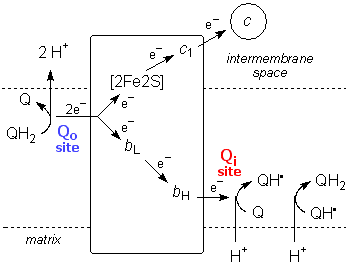
Complex III - Cytochrome bc1 Complex
Each half of dimeric Complex III has a binding site for the lipid-mobile carrier ubiquinol/ubiquinone (QH2/Q). Quinol oxidation takes place near the intermembrane space ("outside", the Qo site), while quinone reduction takes place near the matrix side of the membrane ("inside", the Qi site). Half the electrons delivered at Qo pass through the Complex to Qi, while the other half are delivered to the intermembrane-mobile carrier cytochrome c. Because the redox of Q/QH2 also involves the transfer of protons, this process results in the release of protons into the intermembrane space and the uptake of protons from the cell matrix -- in effect, H+ has been "transported" from one side of the membrane to the other via Q, even though they aren't actually the same protons coming and going during the passage of any given electron through the protein.
There are two pathways for electrons to pass through Complex III from ubiquinol at Qo. One is via two heme iron centres, cytochromes b562 (or bL, with lower potential) and b566 (or bH, with higher potential), and then to ubiquinone at Qi. The other is via a Rieske iron-sulfur cluster to cytochrome c1, and then to cytochrome c.
The overall reaction can be represented as: 2 QH2 + Q + 2H+("in") + 2 cyt cIII ==> 2 Q + 4H+("out") + QH2 + 2 cyt cII
The complete cytochrome bc1 complex consists of a dimer of two identical sets of proteins. Each half is made up of eleven protein subunits and contains the full set of four metal centres. |
In this close up of one half of the complex, two molecules of ubiquinone are seen to either side of the two b hemes, representing one half of the bifurcated electron transfer chain: from Qo (above), through b562 and b566, to Qi, below. Each iron ion is octahedral, with two axial hisN groups. |
| Structure of Complex III from S. cerevisiae (yeast), from Lange, et al. Proc. Natl. Acad. Sci. USA 2002, 99, 2800. PDB 1KYO. | |
In this view, all four metal clusters are seen. Stigmatellin (a bc1 inhibitor, shown in green) occupies the Qo binding site, proximal to both a cytochrome b and the Rieske FeS cluster. The two cytochrome b centres are bound in the yellow protein chains. The second electron transfer path passes through the iron-sulfur cluster and the cytochrome c1 centre (blue chain), delivering electrons to cytochrome c, which binds to the protein outside the mitochondrial membrane (purple chain). Note the ligands to the iron cluster: one iron is bound by two cysS groups, but 2 hisN support the other, giving the cluster a much higher redox potential than normal [2Fe2S] ferredoxin clusters. |
Cytochrome c
|
Structure of cytochrome c from T. alalunga (tuna). PDB 5CYT. |
Cytochrome c is a mobile electron carrier outside the mitochondrial membrane, shuttling one electron at a time from Complex III to Complex IV. It contains a heme group with two axial ligands, one hisN and one metS, giving octahedral coordination to the iron atom. The two iron oxidation states are Fe(III) d5 and Fe(II) d6, both low spin. The transfer of a non-bonding t2g electron doesn't dramatically affect the coordination geometry of the iron atom, making for kinetically facile electron transfer. |
Complex IV - Cytochrome c Oxidase
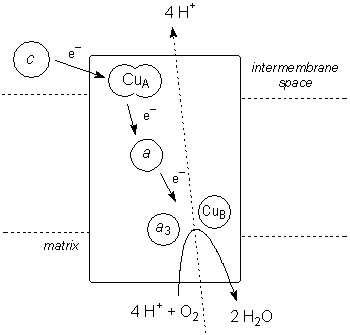
Cytochrome c Oxidase (CcO, Complex IV) is a large, membrane-bound dimeric enzyme, with each half of the dimer consisting of 13 protein chains. The complex acts as the terminus of mitochondrial electron transport in all aerobic life, by using four electrons to reduce dioxygen: O2 + 4H+ + 4e– ==> 2 H2O. This reaction is coupled with the transfer of four protons across the mitochondrial membrane, driving the synthesis of ATP.
In each half of CcO, there are three metal sites involved in electron transfer: bimetallic CuA, monometallic cytochrome a, and bimetallic cytochrome a3/CuB. (In addition, there are non-redox Mg2+ (red) and Zn2+ (green) ions, which, along with the CuB/cyt a3 site, may be involved in the proton-pumping mechanism.) Cytochrome c initiates electron flow through the enzyme by delivering one electron at a time to CuA, which sits just outside the membrane. Electrons flow from CuA through cyt a to the a3/CuB site, which is where O2 is bound and reduced to water. (Note that with four one-electron redox centres — CuA, cyt a, CuB, and cyt a3 — the enzyme can hold the four electrons needed for the complete reduction of a single O2 molecule.)
|
Structure of Complex IV from B. taurus (cow), from Yoshikawa, et al. Science 1998, 280, 1723. PDB 1OCR. |
 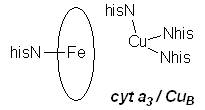
|
|
|
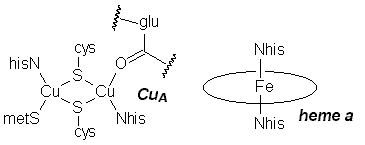
In CuA, one Cu atom is bound by NHis, SMet, and two SCys that bridge the two Cu centres; the other is bound by the two bridging SCys, one NHis, and an unusual carbonyl oxygen from a glutamate residue. In the oxidized form, this site consists of two antiferromagnetically coupled Cu(II) d9 centres, while the reduced form is a delocalized d9/d10 system (in effect, two Cu with a +1.5 oxidation state). Electrons are transferred about 19.5Å from CuA to cytochrome a, a typical heme iron centre with two NHis groups completing octahedral coordination about the metal. The redox couple involves LS d5 Fe(III) and LS d6 Fe(II). Electrons then move 13.5Å from cyt a to the cytochrome a3/CuB site. In the fully reduced form, the iron is CN=5 HS d6 Fe(II), with one NHis group, while the copper is CN=3 d10 Cu(I), bound by three NHis. The two metals are separated by about 5Å. O2 coordinates to this site, bridging between the two metal centres.
|
|
Energetics of the Mitochondrial Electron Transport Chain
Neglecting Complex II, the overall reaction of the mitochondrial chain, per 2e– transferred, can be written as:NADH + H+ + ½ O2 + 10 H+("in") ===> NAD+ + H2O + 10 H+("out") E° = +1.135V
Each two e– through the chain results in the net transfer of 10 protons across the membrane: 4 at Complex I, 4 at Complex III, and 2 at Complex IV. In turn, this proton gradient provides enough energy to synthesize about 2.5 molecules of ATP:
2.5 ADP + 2.5 Pi ===> 2.5 ATP + 2.5 H2O ΔG° = +76 kJ/mol
The total electrochemical potential for this reaction is E° = +1.135V or ΔG° = –219 kJ/mol O2. About 200 kJ of that energy are conserved in the proton gradient, of which about 76 kJ/mol is ultimately stored in ATP, resulting in a total energy efficiency of about 35%.
This page is maintained and copyright by W. Stephen McNeil at UBC Okanagan.
All educational works available on this page are licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 2.5 Canada License. ![]()